
Data Coverage
data_coverage.RmdIntroduction
The data_coverage() function facilitates commonly
performed data coverage checks and is meant as a starting
point for data coverage exploration.
Although GEMINI aims to achieve high coverage of all data elements, it’s possible that some tables could not be fully extracted from certain hospitals/time periods. This is especially true for clinical variables, such as lab, transfusion, radiology, or pharmacy data.
Therefore, one of the first steps of any analysis with GEMINI data should be to carefully check data coverage for all relevant tables. Depending on the research question, this may inform study feasibility, cohort creation, decisions about imputation of missing values, and/or modelling choices.
Expand this section for an example
If the goal of a project is to compare patients who received a blood
transfusion vs. those who did not, researchers first need to check data
coverage for the transfusion table. In this scenario, users cannot
simply check whether a given genc_id has an entry in the
transfusion table because there are 2 possible reasons for why a
genc_id might not exist in the transfusion table:
- Clinical reason: A transfusion may not have been indicated (i.e., the patient did indeed not receive a transfusion).
-
Data coverage issue: The patient did
receive a blood transfusion, but the GEMINI transfusion table does not
contain the corresponding
genc_iddue to data coverage issues (e.g., data extraction issue).
We need to be able to differentiate between these 2 scenarios to distinguish:
-
True negatives: I.e., patient did indeed
not receive a transfusion, so we can set
transfusedtoFALSE. -
Transfusion status unknown: I.e., we do
not have sufficient data coverage to know whether the patient received a
transfusion, so we should set
transfusedtoNA(or exclude patients without transfusion coverage from the cohort).
Additionally, if the goal is to analyze differences in transfusion practices across hospitals, we also need to check whether there are differences in data coverage timelines across sites. For example, if some hospitals only have transfusion data before 2020, whereas other hospitals only have transfusion data after 2020, any between-hospital differences we observe may in fact be driven by changes in transfusion practices over time (rather than reflecting true differences between hospitals). We therefore need to carefully consider which hospitals & time periods to include in our analyses, and how to account for differences in data coverage periods.
This vignette focuses on data coverage checks at the level of
tables - not individual columns within a given table.
This is because table-level data coverage is typically more challenging
to analyze and requires a thorough inspection and understanding of the
GEMINI cohort. By contrast, column-level missingness within a single
table can easily be checked using Rgemini::n_missing().
Column-level missingness should typically follow the table-level
coverage checks discussed here.
Data coverage lookup table
The data_coverage() function queries the
lookup_data_coverage table, which lists the time periods
with coverage for each data table and hospital (also see Data
Availability plot in the Data
Dictionary). The min/max dates refer to the range of
discharge dates (from admdad) during which
there was at least 1 genc_id with an entry
in a given table:
lookup_data_coverage <- dbGetQuery(
db, "SELECT * from lookup_data_coverage;"
) %>% data.table()This is an example (dummy version) of the
lookup_data_coveragetable showing the min-max coverage dates for different hospital*data combinations:
| data | hospital_num | min_date | max_date | additional_info |
|---|---|---|---|---|
| admdad | 1 | 2018-02-01 | 2023-03-31 | Hospital opened in February 2018. |
| er | 2 | 2015-04-01 | 2023-06-30 | |
| pharmacy | 3 | 2019-07-01 | 2023-03-31 | |
| radiology | 4 | 2017-01-01 | 2023-06-30 | |
| transfusion | 5 | 2015-04-01 | 2022-05-31 | |
| ipscu | 6 | NA | NA | Hospital does not have an ICU. |
| ipdiagnosis | 7 | 2015-04-01 | 2023-06-30 | |
| lab | 8 | 2017-04-01 | 2018-04-07 | Hospital provides specialized care. |
| lab | 8 | 2018-12-10 | 2021-10-15 | Hospital provides specialized care. |
| lab | 8 | 2022-03-30 | 2023-03-20 | Hospital provides specialized care. |
| physicians | 9 | 2016-04-01 | 2023-03-31 |
Additional notes:
- The min/max dates in each row correspond to the start/end date of a single data coverage period by hospital*table.
- Hospital*table combinations with interrupted timelines are shown across multiple rows (e.g., 3 rows for lab data at site 8).
- Gaps in data coverage are defined as a period of >28 consecutive days without any data.
- Rows where the dates are
NAindicate hospital*table combinations for which GEMINI does not have any data at all. This could either mean that the data are not relevant to a given site (e.g., hospital 6 does not have an ICU) or that the data could not be successfully extracted. - Encounters with a discharge date within the listed min-max periods are assumed to have at least some data coverage for that table, although coverage may still be low (see plot_coverage below).
- The
additional_infocolumn contains information that may be helpful when interpreting the output of thedata_coverage()function. When runningdata_coverage(), the function automatically prints all relevant rows from this column in the terminal.
WARNING: Our threshold
for data coverage is currently extremely low! We assume that
there is some data coverage if there is at
least 1 genc_id with an entry in a given table.
This does not tell us how high data coverage actually is (e.g., was
there only 1 genc_id or 1,000 genc_ids with an
entry in a given table?). Additionally, the definition only considers
gaps >28 days, and does not exclude time periods with shorter gaps.
Therefore, users should only use this lookup table (and the accompanying
data_coverage() function) as a starting point - to exclude
time periods where GEMINI did not receive any data at all. We highly
recommend performing additional coverage checks, in combination with
data quality assessments.
How to use data_coverage()
Get encounter-level coverage flag (without plots)
The data_coverage() function converts the
lookup_data_coverage table to an encounter-level table.
Let’s say we want to check which encounters in admdad were
discharged during time periods with any lab,
transfusion, and radiology data coverage:
# Get encounter-level coverage flags
coverage <- data_coverage(
dbcon = db,
cohort = admdad,
table = c("lab", "transfusion", "radiology"),
plot_timeline = FALSE,
plot_coverage = FALSE
)This is an example (dummy version) of the ouput table returned by
data_coverage():
| genc_id | hospital_num | discharge_date | lab | transfusion | radiology |
|---|---|---|---|---|---|
| 1 | 1 | 2019-12-31 | TRUE | FALSE | FALSE |
| 2 | 2 | 2022-03-12 | TRUE | TRUE | TRUE |
| 3 | 3 | 2018-10-10 | TRUE | TRUE | TRUE |
| 4 | 4 | 2017-10-15 | FALSE | TRUE | TRUE |
| 5 | 5 | 2022-06-01 | FALSE | FALSE | TRUE |
| 6 | 6 | 2018-12-22 | TRUE | FALSE | TRUE |
| 7 | 7 | 2022-05-20 | TRUE | TRUE | TRUE |
| 8 | 8 | 2021-07-30 | TRUE | FALSE | TRUE |
| 9 | 9 | 2017-08-03 | FALSE | TRUE | TRUE |
The table contains a flag for each genc_id indicating
whether that genc_id was discharged from a hospital &
during a time period where some
lab/transfusion/radiology data were available (according to the min-max
dates in the lookup_data_coverage
table).
-
For
genc_idswhere the flag isFALSE: Based on the definition of data coverage in the lookup table, these are encounters that were discharged from hospitals/during time periods where GEMINI did not receive ANY data all for a period of >28 days. In other words, we do not have sufficient data coverage to infer whether the encounter received a lab/imaging test or transfusion. Therefore, thesegenc_idsshould be excluded (or set toNA) for any analyses relying on lab/transfusion/radiology data. -
For
genc_idswhere the flag isTRUE: These are encounters that were discharged from hospitals/during time periods with some data coverage. This does not mean that allgenc_idswhere the flag isTRUEhave an entry in the lab/transfusion/radiology table because:- Due to the low threshold for data coverage in the
lookup_data_coveragetable, actual data coverage may still be low during some time periods (seeplot_coveragebelow). - For some tables, we would not expect all
genc_idsto have an entry even if data coverage is generally high (e.g., not all patients receive a radiology test or blood transfusion).
- Due to the low threshold for data coverage in the
Plot data timelines by table & hospital
To visualize the data timelines by hospital & table, you can set
the plot_timeline input to TRUE, which will
result in a plot similar to the “Data Availability” plot in the data
dictionary. However, the plot returned by this function is customized to
the input cohort (e.g., time periods/hospitals of interest) and specific
tables that are relevant for your analyses.
For example, let’s say we are interested in the data timelines for
the admdad, lab, and ipscu table
for encounters discharged since April 2017:
# Get encounters discharged since Apr 2017
admdad_subset <- admdad[discharge_date_time >= "2017-04-01 00:00", ]
# Plot data timelines
coverage <- data_coverage(
dbcon = db,
cohort = admdad_subset,
table = c("admdad", "lab", "ipscu"),
plot_timeline = TRUE,
plot_coverage = FALSE
)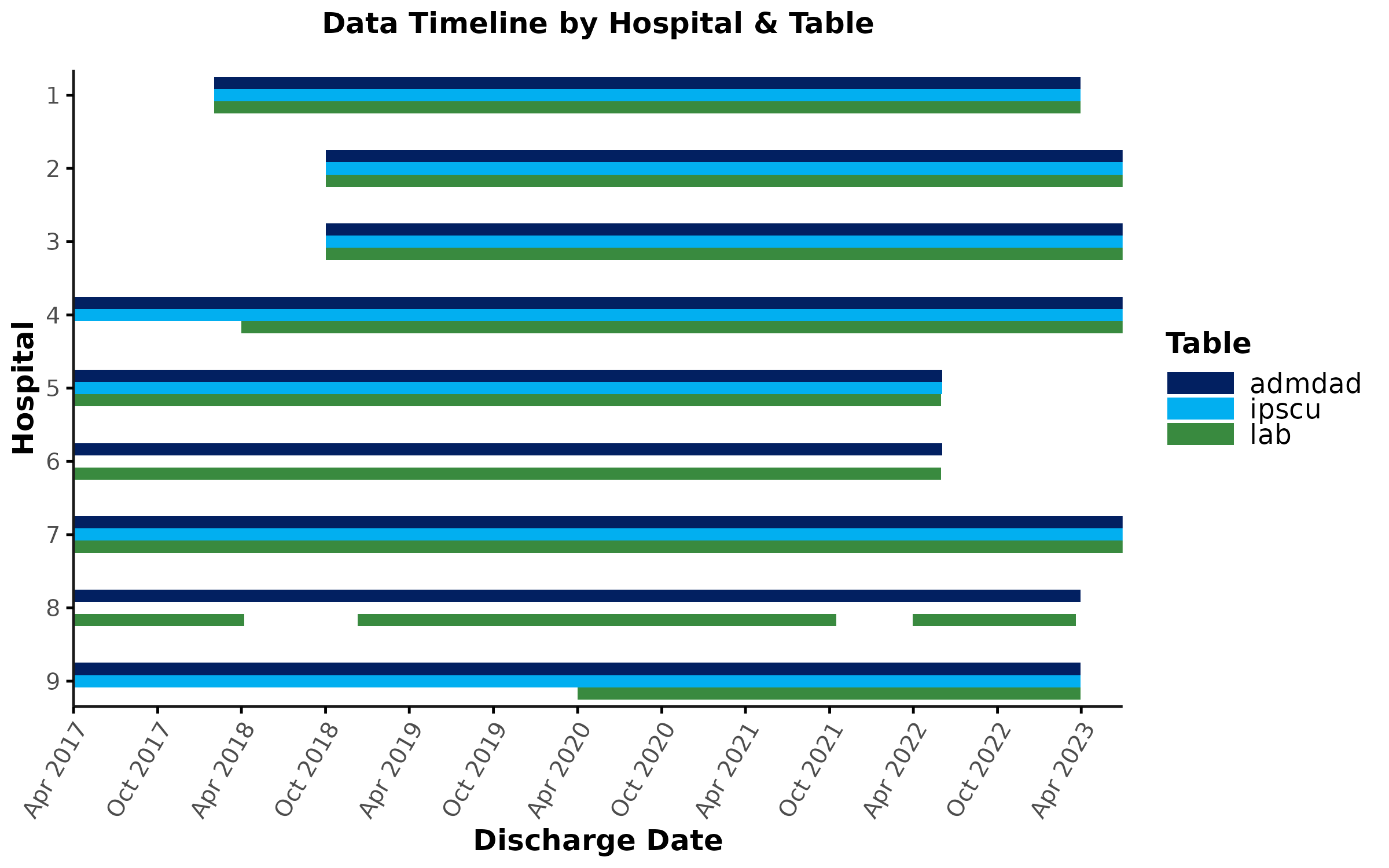
Let’s inspect this plot. What should you look out for?
Expand this section for some tips
-
Between-hospital differences in data timelines
- Looking at the timelines for
admdad, you’ll notice that some hospitals generally have later start dates (e.g., hospital 1) and/or earlier end dates (e.g., hospitals 5 & 6) compared to others. Theadditional_infocolumn in thelookup_data_coveragetable may provide an explanation for some of these between-hospital differences in data timelines (e.g., see above, for hospital 1: “Hospital opened in February 2018.”). Depending on your research question, you may need to restrict your analyses to time periods that overlap across all sites and/or exclude sites with limited data timelines.
- Looking at the timelines for
-
Within-hospital differences in data timelines
- Even within the same hospital, data timelines can vary between
tables. For example at sites 4 and 9, lab data have a shorter coverage
period compared to
admdad/labdata. This is typically due to challenges associated with extracting clinical (as opposed to administrative) data. If lab data are crucial for your research question, keep in mind that the effective data timeline for your analyses is determined by lab data coverage (rather thanadmdadcoverage).
- Even within the same hospital, data timelines can vary between
tables. For example at sites 4 and 9, lab data have a shorter coverage
period compared to
-
Interrupted data timelines
- For some hospitals/tables, data timelines may be interrupted. For
example, at site 8, lab data are not covered continuously. The
additional_infocolumn may contain some information that can help interpretation. For example, for site 8 lab data, the table states that this “Hospital provides specialized care”. It’s possible that lab testing is not performed on a routine basis at specialized hospitals and that the % of encounters with lab tests is generally very low even for time periods that are shown as “covered” in the timeline plot (remember that a singlegenc_idwith lab data is sufficient for data coverage to be shown asTRUEin this plot). In other words, the gaps in the data timeline might not necessarily reflect a data extraction issue, but rather could be an indicator of specialized clinical practice at this site. Therefore, it is important to inspect overall data volume for sites with interrupted data timelines and plot the % data coverage (see plot_coverage below). If lab data are crucial for your research question, you may consider exlcuding specialized hospitals with low lab data coverage/volume.
- For some hospitals/tables, data timelines may be interrupted. For
example, at site 8, lab data are not covered continuously. The
-
Empty rows
- Some tables may not appear in the timeline plot at all for certain
hospitals. For example, sites 6 & 8 have an empty
ipscurow. According to theadditional_infocolumn in thelookup_data_coveragetable, site 6 does not have an ICU. This information can help us determine how to deal with this site in our analyses. For example, if we are simply looking at theipscutable to determine if agenc_idwas admitted to an ICU during their hospitalization, we can still include site 6 in our analyses and seticu_entrytoFALSEfor all encounters at this site (we know for a fact that patients were not admitted to an ICU while being hospitalized at site 6). By contrast, if we are planning to perform more in-depth analyses ofipscudata (e.g., analyses of lab tests performed in the ICU), site 6 would need to be excluded since it would not contribute any data points to those analyses.
- Some tables may not appear in the timeline plot at all for certain
hospitals. For example, sites 6 & 8 have an empty
In addition to the plot, the function also returns a list object with several entries:
- The 1st list item (
coverage_flag_enc) contains the table with encounter-level flags based onlookup_data_coverage(also see section above):
coverage[[1]] # = coverage$coverage_flag_enc| genc_id | hospital_num | discharge_date | admdad | ipscu | lab |
|---|---|---|---|---|---|
| 1 | 1 | 2018-09-13 | TRUE | TRUE | TRUE |
| 2 | 7 | 2023-02-01 | TRUE | TRUE | TRUE |
| 3 | 7 | 2023-01-20 | TRUE | TRUE | TRUE |
| 4 | 1 | 2018-07-10 | TRUE | TRUE | TRUE |
| 5 | 9 | 2019-10-10 | TRUE | TRUE | FALSE |
| 6 | 4 | 2019-09-21 | TRUE | TRUE | TRUE |
| 7 | 9 | 2020-12-26 | TRUE | TRUE | TRUE |
| 8 | 4 | 2020-12-06 | TRUE | TRUE | TRUE |
| 9 | 4 | 2019-04-23 | TRUE | TRUE | TRUE |
| 10 | 9 | 2022-03-02 | TRUE | TRUE | TRUE |
- The 2nd list item (
timeline_data) is the DB availability table with min-max dates per site & table (i.e., what’s shown in the data timeline plot)
coverage[[2]] # = coverage$timeline_data| hospital_num | data | min_date | max_date | additional_info |
|---|---|---|---|---|
| 1 | admdad | 2018-02-01 | 2023-03-31 | NA |
| 1 | ipscu | 2018-02-01 | 2023-03-31 | NA |
| 1 | lab | 2018-02-01 | 2023-03-31 | NA |
| 2 | admdad | 2018-10-01 | 2023-06-30 | NA |
| 2 | ipscu | 2018-10-01 | 2023-06-30 | NA |
| 2 | lab | 2018-10-01 | 2023-06-30 | NA |
| 3 | admdad | 2018-10-01 | 2023-06-30 | NA |
| 3 | ipscu | 2018-10-01 | 2023-06-30 | NA |
| 3 | lab | 2018-10-01 | 2023-06-30 | NA |
| 4 | admdad | 2017-04-01 | 2023-06-30 | NA |
-
The 3rd list item (
timeline_plot) is a ggplot object containing the timeline plot, which can be further customized/exported in the desired format.For example, you could customize the title of the plot and move the legend position:
coverage[[3]] + # = coverage$timeline_plot
ggtitle("Data coverage exploration\n\n", subtitle = "GEMINI project XYZ") +
theme(legend.position = "top")Plot % data coverage
The encounter-level flags and timeline plot only provide very limited
insights into actual data coverage, since they simply reflect the
min-max dates listed in the lookup_data_coverage table,
which lists all time periods with any data even if just 1
genc_id had a table entry. This is problematic because
there may be time periods with very low coverage that aren’t currently
excluded from the lookup_data_coverage table. Therefore,
users should always perform additional coverage checks by plotting the %
of encounters with an entry in a given table by hospital & month.
This can be achieved by setting the plot_coverage input to
TRUE. Users should then carefully inspect the coverage over
time and decide how to handle time periods/hospitals with low coverage
in their analyses.
WARNING: When plotting data coverage, your cohort input should only be pre-filtered for the hospitals and overall timelines of interest. You should not apply any other cohort inclusion/exclusion criteria (e.g., diagnosis codes etc.). This is to ensure that the coverage plots are representative of the overall GEMINI data holdings, and are not skewed by any project-specific cohort inclusion/exclusion steps.
Let’s plot coverage for the transfusion table for patients discharged since April 2017:
# Plot coverage (% genc_ids with entry in table)
coverage <- data_coverage(
dbcon = db,
cohort = admdad_subset,
table = c("transfusion"),
plot_timeline = FALSE,
plot_coverage = TRUE
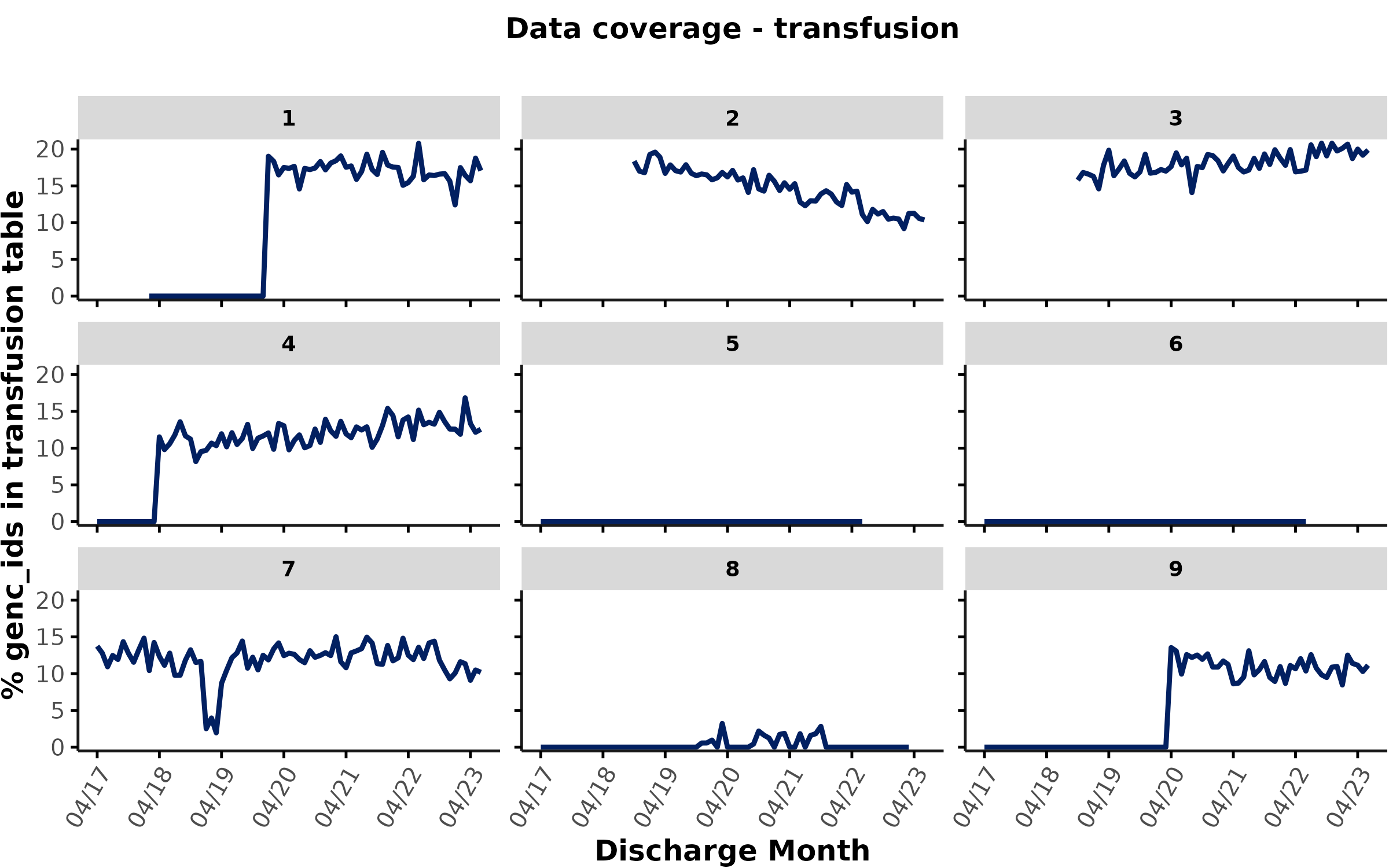
)
The expected percentage of genc_ids with an entry in a
given table depends on the clinical context & differences in
practice across sites (e.g., how many patients receive a transfusion
vs. a lab/imaging test etc.). Therefore, there is no single threshold
that can be applied to define “good” coverage across different
tables/hospitals.
With that in mind, how would you evaluate this coverage plot for transfusion data?
Expand this section to learn more
Users should carefully inspect these plots for unexpected patterns (e.g., sudden drops) and further inspect a) what’s driving these patterns, and b) how to handle time periods with low data coverage in their analyses. Here are a few specific examples of what to look out for:
-
Hospitals/time periods with no data coverage
- Some hospitals don’t have any transfusion data at all (e.g., sites 5
& 6) or don’t have any transfusion coverage during certain time
periods (e.g., hospital 1/4/8/9).* Note that time periods with 0%
coverage could also be inferred from the timeline plot, which would have
empty rows for any tables without any data coverage (see
plot_timeline). However, the coverage plots provide more granular insights into coverage - beyond 0 vs. non-0 - allowing users to compare coverage between hospitals and over time on a continuous scale.
- Some hospitals don’t have any transfusion data at all (e.g., sites 5
& 6) or don’t have any transfusion coverage during certain time
periods (e.g., hospital 1/4/8/9).* Note that time periods with 0%
coverage could also be inferred from the timeline plot, which would have
empty rows for any tables without any data coverage (see
-
Differences between hospitals
- For hospitals with transfusion data, we may see large differences in
the % of encounters with an entry in the transfusion table (e.g.,
>15% at site 3 vs. <5% at site 8). This may not necessarily
reflect differences in data coverage, but could be due to differences in
clinical practice or medical subservices provided at different sites. As
discussed above, we already know that site 8 provides specialized care.
To check whether this might explain low transfusion rates, users could
inspect the medical subservices provided at this site (e.g.,
mrp_servicefield inadmdad) and could also check for potential differences in patient populations (e.g., most responsible diagnosis groups) at different sites. If transfusion data are crucial for the research question of interest, it may make sense to exclude hospitals with low transfusion data volume.
- For hospitals with transfusion data, we may see large differences in
the % of encounters with an entry in the transfusion table (e.g.,
>15% at site 3 vs. <5% at site 8). This may not necessarily
reflect differences in data coverage, but could be due to differences in
clinical practice or medical subservices provided at different sites. As
discussed above, we already know that site 8 provides specialized care.
To check whether this might explain low transfusion rates, users could
inspect the medical subservices provided at this site (e.g.,
-
Sudden drops in data coverage
- At hospital 7, we can see a large drop in the % transfusion coverage in the first quarter of 2019. Sudden, temporary drops like this typically reflect a data coverage issue (e.g., due to data extraction challenges, EHR transition etc.). Occassionally, these drops could also be due to temporary changes in clinical practice (e.g., COVID-19 waves, blood product shortages etc.) - although we would typically expect these changes to be more gradual and affect all hospitals. Depending on your research question, you may need to remove hospitals/time periods with low data coverage from your analyses, especially if you suspect a data coverage issue. Note that coverage drops like this would not be reflected in the timeline plot shown above because coverage was > 0%.
-
Gradual changes in coverage over time
- At site 2, we can observe a gradual decrease in the % of encounters
with a transfusion. This could be due to several different reasons:
- There may have been a change in clinical practice over time (e.g., new transfusion guidelines specific to this site)
- Changes in the patient cohort/medical subspecialties: It’s worth inspecting if patient characteristics remained constant over time, or whether there are reasons you’d expect transfusion rates to decrease over time (e.g., larger proportion of patients with no medical indication for a transfusion?)
- It’s possible that gradual changes in coverage are driven by data
extraction issues. For example, later time periods may be missing
transfusion entries for a certain subset of the GEMINI cohort (e.g., ICU
patients). Hence, it could be worth plotting coverage for different
patient cohorts separately (see cohort flags like
gimorall_medin thederived_variablestable). - Finally, note that we are plotting data coverage by discharge month here (rather than transfusion month). This can potentially mask issues related to the extraction of transfusion products that were transfused during certain time periods. We therefore recommend plotting coverage by the relevant clinical date-time variable (i.e., issue_date_time for transfusions) in addition to the plots by discharge_date_time. Note that this is currently not supported by data_coverage() but can easily be achieved using Rgemini::plot_over_time() (e.g., time_var would be issue_date_time in the case of transfusions). This may show a more abrupt drop in data coverage and could indicate issues with data storage in the hospital’s EHR (e.g., system outage) rather than data extraction issues (which are typically related to discharge_date_time since GEMINI data are pulled by discharge date).
- At site 2, we can observe a gradual decrease in the % of encounters
with a transfusion. This could be due to several different reasons:
admdad data are available), but none of the encounters had
an entry in the transfusion table. By contrast, time
periods that are not covered in the cohort/admdad table are shown as
NA in these plots. For example, at site 1, we can see that
the cohort only starts in February 2018 in line with the
additional_info column stating that this hospital only
opened in February 2018. between Feb 2018 - Jan 2020, GEMINI did not
receive any transfusion data for the encounters from this site (%
coverage = 0). Since January 2020, GEMINI has started receiving
transfusion data with ~15-20% of encounters having an entry in the
transfusion table at this site.
In addition to printing the coverage plot, the function will also return a list object similar to the one above:
-
coverage_flag_enc: Encounter-level data coverage flag -
coverage_data: Table showing the % of encounters with a table entry by hospital & discharge month -
coverage_plot: ggplot object containing coverage plot
Here is an example of the coverage_data table, which can
be used to detect (and exclude) hospitals/months with low coverage. It
also shows the number of encounters (n_encounters) that are
in the denominator for each hospital * month combination based on the
user-provided cohort input.
coverage[[2]] # = coverage$coverage_data| discharge_month | hospital_num | prct_transfusion_entry | n_encounters |
|---|---|---|---|
| 2017-04-01 | 1 | NA | NA |
| 2017-04-01 | 2 | NA | NA |
| 2017-04-01 | 3 | NA | NA |
| 2017-04-01 | 4 | 0.00 | 533 |
| 2017-04-01 | 5 | 0.00 | 1145 |
| 2017-04-01 | 6 | 0.00 | 700 |
| 2017-04-01 | 7 | 13.74 | 844 |
| 2017-04-01 | 8 | 0.00 | 1 |
| 2017-04-01 | 9 | 0.00 | 652 |
| 2017-05-01 | 1 | NA | NA |
Further customization
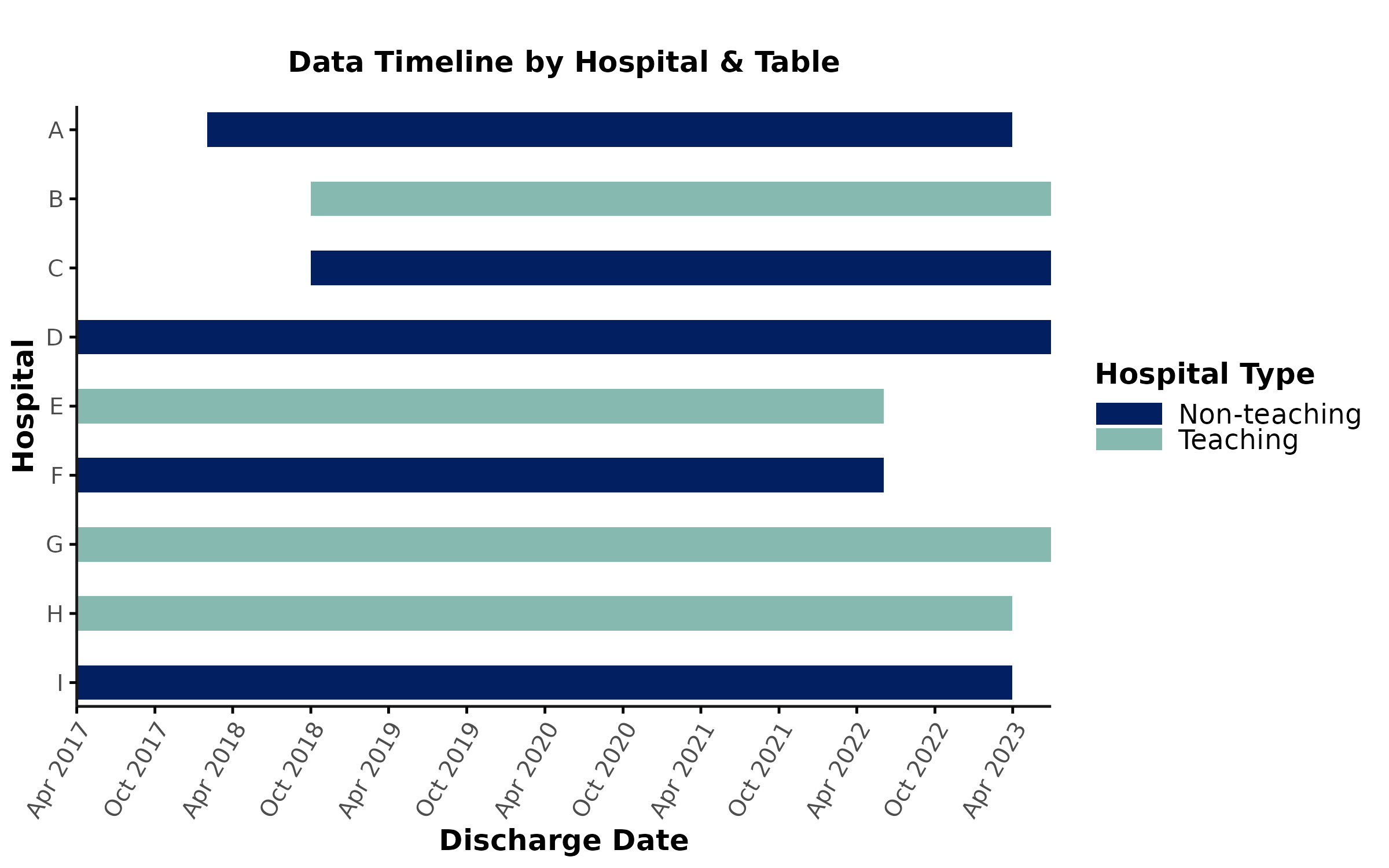
hospital_label & hospital_group
Users can further customize the timeline & coverage plots by
providing hospital_label (e.g., letters A-E instead of
hospital_num 101-105) and/or hospital_group
(e.g., “Teaching” vs. “Non-teaching hospital”) inputs.
For example, let’s say our admdad_subset table contains
a user-created label (hosp_label) and hospital grouping
variable (hosp_type). We can now provide these variable
names as inputs to data_coverage:
hospital_label will change the labels of hospitals on the
y-axis of the timeline plot whereas hospital_group will
color code hospitals according to their category. We can also customize
the color scheme by providing a colors input:
coverage <- data_coverage(
dbcon = db,
cohort = admdad_subset,
table = c("admdad"),
plot_timeline = TRUE,
plot_coverage = FALSE,
hospital_label = "hosp_label",
hospital_group = "hosp_type",
colors = gemini_colors(3)
)
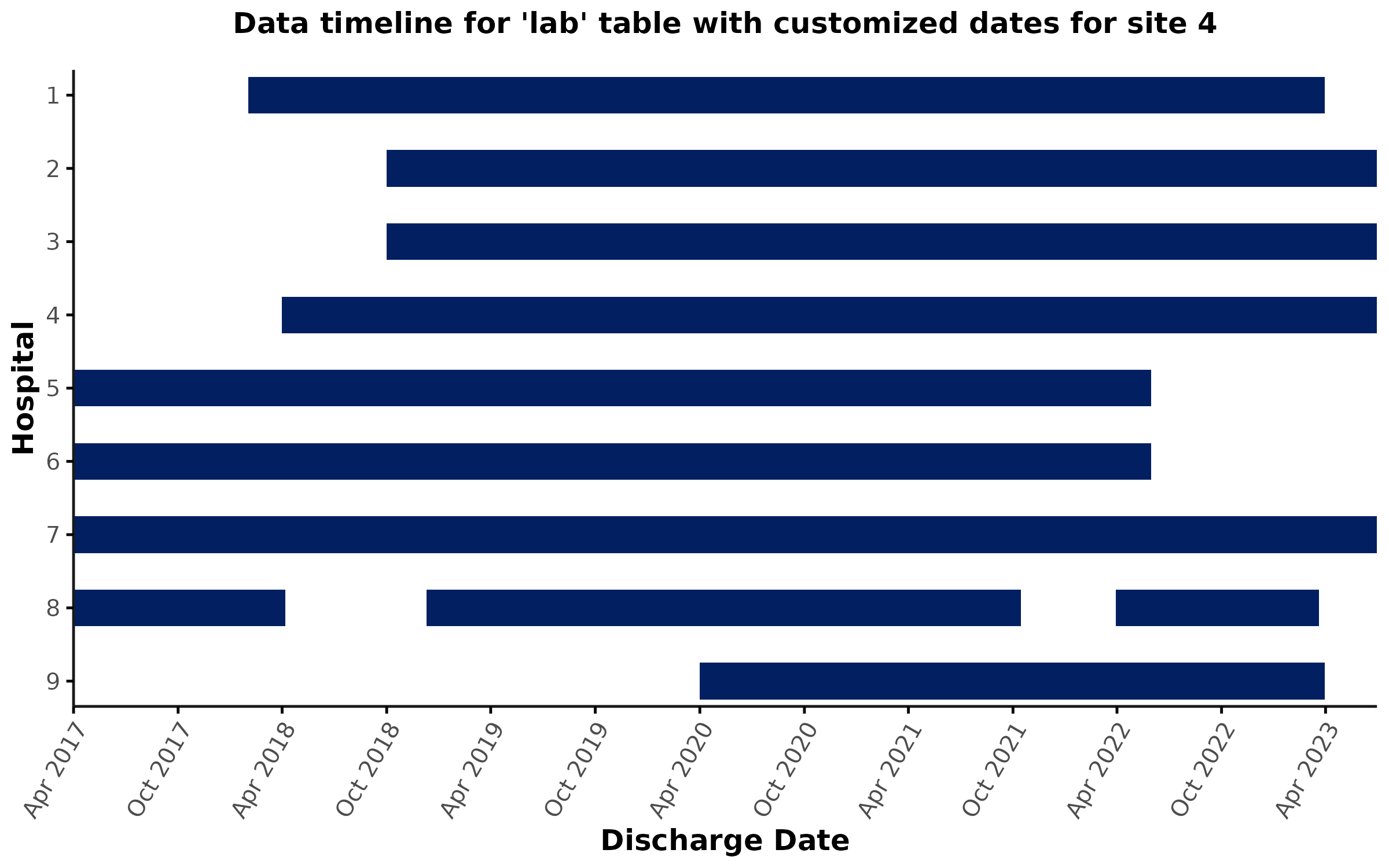
custom_dates
In some cases, users may want to provide customized timelines for a
given hospital*table combination to apply further inclusions/exclusions
(e.g., due to data quality issues, column-level missingness, etc.). For
example, let’s say you identified a data quality issue with lab data at
hospital 4 for discharge dates prior to April 2019 and now only want to
include encounters that were discharged after that date. This can be
achieved by providing a custom_dates input to
data_coverage(), which will overwrite the rows for the
corresponding hospital*table combinations in the
lookup_data_coverage table. As a result, the
encounter-level flags and timeline plot will be adjusted according to
the user-specified dates. Note that the coverage plot (see above) is not affected by the
custom_dates input.
# define customized dates
custom_dates <- data.table(
data = c("lab"),
hospital_num = c(4),
min_date = "2019-04-01",
max_date = "2023-06-30"
)
coverage <- data_coverage(
db = dbcon,
cohort = admdad_subset,
table = c("lab"),
plot_timeline = TRUE,
plot_coverage = FALSE,
custom_dates = custom_dates
)
as_plotly
Finally, users can set as_plotly to TRUE.
This will return any figures created by data_coverage() as
ggplotly objects, which facilitates interactive exploration
of the timeline/coverage plots. Note that this is currently not
supported on HPC4Health due to vulnerabilities associated with the
plotly package.