
Episodes of care and Readmission
epicare_and_readmission.RmdIntroduction
This vignette contains detailed information about two functions that
are closely related to each other: episodes_of_care() and
readmission(). The episodes_of_care function
is used in the readmission calculations in order to avoid
that encounters that are linked via transfers (i.e., are part of the
same episode of care) are counted as readmissions. The definitions used
in both functions are based on CIHI guidelines for readmission
calculations (see here).
episodes_of_care()
The function episodes_of_care() links
together hospitalizations with acute care transfers into unique episodes
of care. For each hospitalization (genc_id), the function
returns the corresponding: patient_id_hashed,
time_to_next_admission,
time_since_last_admission, AT_in_coded,
AT_out_coded, AT_in_occurred,
AT_out_occurred, epicare.
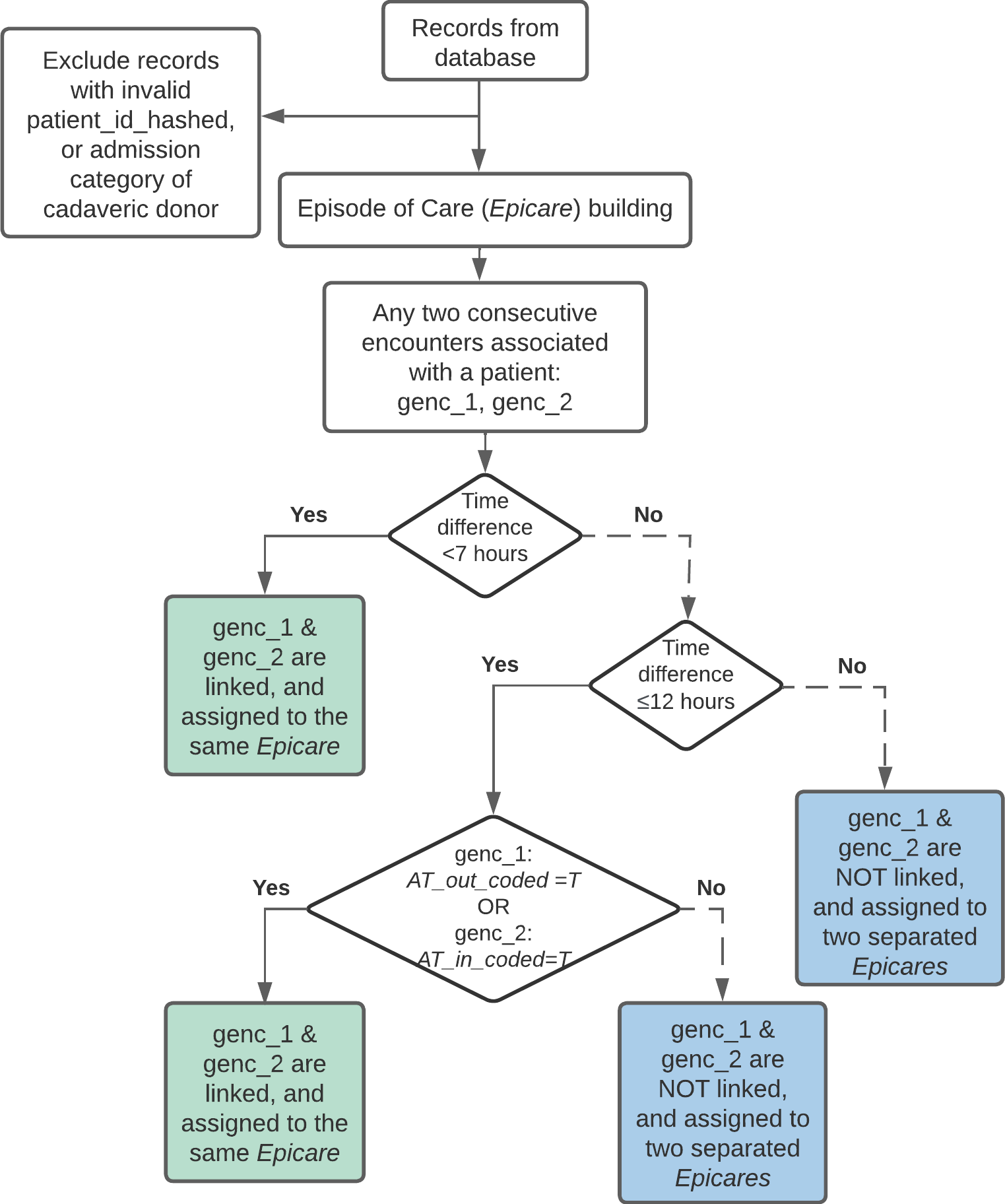
Coded Transfers (AT_in_coded, AT_out_coded): A coded acute care transfer refers to hospital coding of acute care transfers based on DAD
institution_fromandinstitution_tofields. Specifically, a hospitalization has a coded transfer-in (AT_in_coded = T), if thelookup_transferstable listsinstitution_from_type_mns == 'AT'for a givengenc_id(andAT_out_coded = Tifinstitution_to_type_mns == 'AT').Occurred Transfers (AT_in_occurred, AT_out_occurred): An acute care transfer is assumed to have occurred (
AT_in_occurred = T,AT_out_occurred = T) if either of the following criteria are met:
- An admission to a GEMINI hospital within 7 hours after discharge from another GEMINI hospital, regardless of whether the transfer is coded by hospital.
- An admission to a GEMINI hospital within 7-12 hours after discharge from another GEMINI hospital, and at least 1 hospital has coded the transfer.
-
Episode of Care (epicare): An episode of
care refers to all contiguous inpatient hospitalizations within the
GEMINI network. Contiguous inpatient hospitalizations are defined based
on occurred transfers (
AT_in_occurred == TorAT_out_occurred == T). Episodes involving inter-facility transfers are linked regardless of diagnosis.
The flowchart below shows how epicares are built:
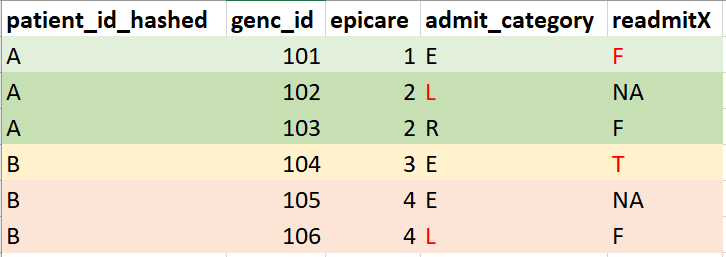
Example output of
episodes_of_care(db)(mock data for illustration purposes, not real data):
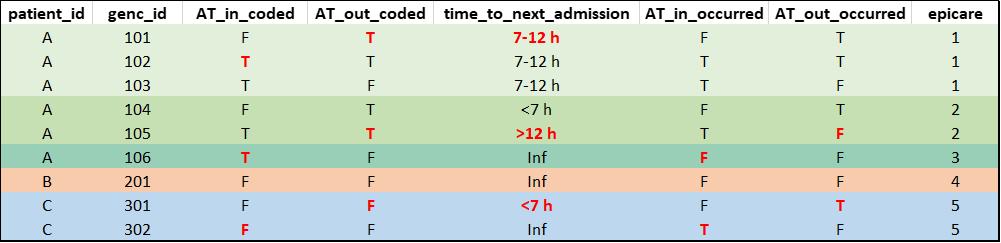
Note: For database versions
drm_cleandb_v4/H4H_template_v5 or newer, the
epicare variable in the derived_variables
table was derived exclusively based on encounters with a medical/special
care unit (SCU) touchpoint since these encounters are consistently
available across hospitals and represent the main patient population of
interest for readmission calculations used in research projects/quality
reports. For all other in-patient encounters (e.g., surgery patients
available at some sites), epicare = NA. See Section 3.4 for options to compute
episodes of care using other, customized cohorts.
readmission()
The function readmission() computes
whether or not a patient associated with an epicare was readmitted to a
GEMINI hospital within a specified time window. For each hospitalization
(genc_id), the function returns its corresponding:
AT_in_occurred, AT_out_occurred,
epicare, and readmitX. readmitX
is a boolean variable denoting whether readmission took place during a
time window X-days post-discharge.
It is important to realize that readmission is determined by looking
forward into data in the future. Therefore, readmitX is a
measurement of quality of care of the current episode and evaluates
whether factors in the current episode LEAD to
readmission in the future.
For episodes of care involving acute care transfers, readmissions are
attributed to the LAST hospitalization from which the
patient was discharged before readmission; namely only the last
genc_id of an episode has readmitX assigned,
the rest are set to NA.
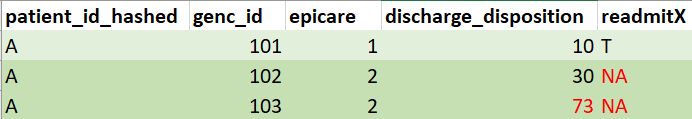
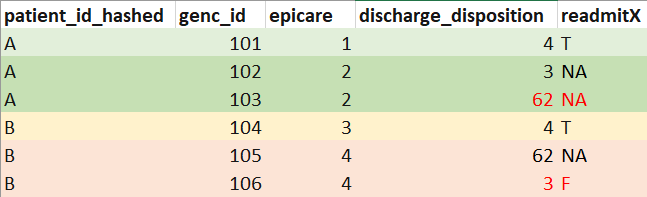
Mock example output of
readmission(db)(mock data for illustration purposes, not real data. patient_id added to facilitate interpretation):
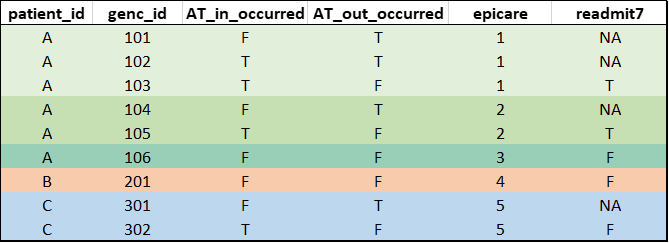
Note: For database versions
drm_cleandb_v4/H4H_template_v5 or newer, the
readmission flags in the derived_variables
table were derived exclusively based on encounters with a medical/SCU
touchpoint since these encounters are consistently available across
hospitals and represent the main patient population of interest for
readmission calculations used in research projects/quality reports. For
all other in-patient encounters (e.g., surgery patients available at
some sites), readmission = NA. See Section 3.4 for options to compute
readmission flags using other, customized cohorts.
General readmission rate calculation
- Denominator of Readmission Rate: Total number of episodes with potential to be readmitted. There are various conditions where episodes are removed from the denominator. For example, episodes discharged as death cannot be readmitted, and are therefore removed from the denominator of readmission rates by default (see details in Section 2 CIHI flags).
An episode is removed from the Denominator by setting
readmitX = NAfor the current episode.
- Numerator of Readmission Rate: Number of episodes within the denominator that were actually readmitted within the time window of interest. There are various conditions where episodes are removed from the numerator. For example, elective admissions might not be considered as readmissions, and are therefore removed from the numerator by default (see details in Section 2 CIHI flags).
An episode is removed from the Numerator by setting
readmitX = Ffor the preceding episode.
Typically, we recommend using the readmission_Xd_derived
variables in the derived_variables table to calculate
readmission rates since these flags were derived using all adult
medicine/SCU encounters available in GEMINI data, and therefore, provide
the most reliable estimates of readmission rates. However, if you want
to calculate readmission flags yourself, the following code illustrates
how to calculate 7-day readmissions using the default arguments in the
readmission() function. Note that this example uses all
encounters available in your database/data cut to derive episodes of
care and subsequent readmissions. This may not be desirable, depending
on your project-specific definition of readmission rates (also see Section 3.4):
# Load necessary libraries:
library(RPostgreSQL)
library(DBI)
library(getPass)
# Establish database connection
db <- DBI::dbConnect(drv,
dbname = "db",
host = "domain_name.ca",
port = 1234,
user = getPass("Username: "),
password = getPass("Password: ")
)
# Run default readmission calculation (with elective_admit = T and death = T)
# this derives epicares/readmission based on all genc_ids in the database/datacut
readm <- readmission(db)
# Compute readmission rate based on readmission output
denominator <- sum(!is.na(readm$readmit7))
numerator <- sum(readm$readmit7, na.rm = T)
readmission_7d_derived <- numerator / denominator
# Compute readmission rate based on readmission output (Alternative)
readmission_7d_derived <- mean(readm$readmit7, na.rm = T)GEMINI specific considerations
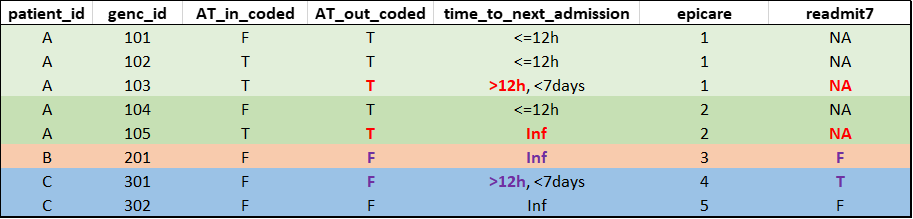
Readmission attribution when end of episode is unknown
When the last hospitalization of an epicare has transfer out coded (
AT_out_coded== T) andtime_to_next_admissionis greater than 12 hours or infinite, it suggests that the patient likely had been transferred to a hospital/service not captured in GEMINI data. It cannot be known for certain that the last hospitalization in our records is the last hospitalization of the epicare in reality.To avoid inaccurate attribution of potential readmission, such epicares are excluded from the denominator (
readmitX = NA).
Hypothetical example of special readmission attribution (compare colored entries in genc_id 103 vs 301, and genc_id 105 vs 201, and see how readmit7 is affected):
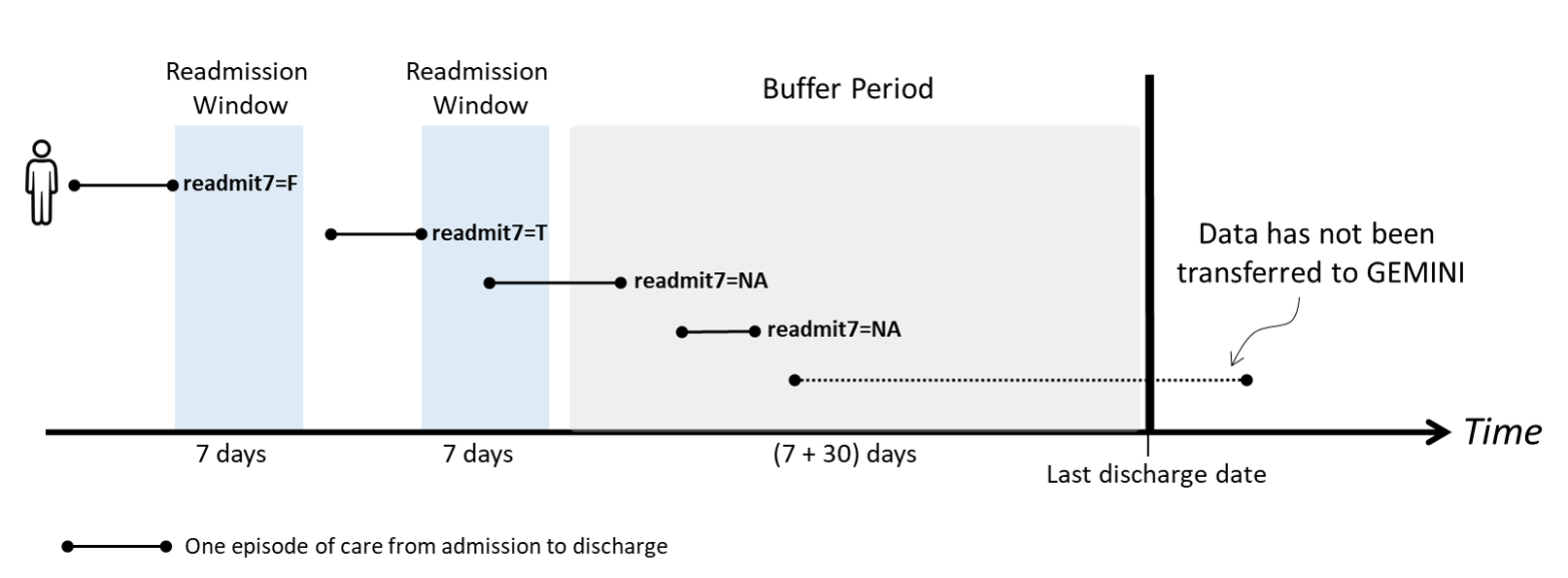
Buffer period based on each hospital’s data availability
A patient needs to be discharged from hospital before appearing in the GEMINI database. If a patient was readmitted to a hospital but has not yet been discharged by the time data is transferred to GEMINI, that readmission record will not be captured in our database. As a result, readmission rates could be underestimated in the time period before the latest data transfers.
To mitigate this bias, a buffer period based on data availability of each hospital is applied to the readmission calculation. If an episode’s discharge time is in the buffer period, the epicare is excluded from the denominator (
readmitX = NA) of readmission rates.
Buffer Period =
[ (Hospital’s last discharge date - Readmission window - Additional 30 days)
, (Hospital’s last discharge date) ]
- The buffer period is derived as follows:
- The X number of days specified by the readmission window must have passed to allow for X-day readmission to occur;
- 95% of hospital admissions are discharged within 30 days based on GEMINI data. Therefore, an additional 30 days are included in the buffer period to exclude any patients who may not yet have been discharged from their readmission encounter;
- The buffer period is determined by the data availability timeline of the hospital where the index admission occurred (since the majority of readmissions occur at the same hospital as the index admission). However, it is possible that there are inter-hospital biases, which cannot be addressed with the current data available in GEMINI.
Hypothetical example of how buffer period affects 7-day readmission of a patient:
Customizable features
CIHI flags: see Section 2 CIHI flags.
Readmission time window: By default, the
readmission()function computes 7-day and 30-day readmission rates. Users have the option to specify a customized time window using thereadm_winargument (see Section 3.3 for details).Restricted cohort: By default, the
readmissionfunction includes all encounters in the database. Depending on your database version and project-specific needs, this may not be desirable. Both theepisodes_of_care()andreadmission()functions therefore allow users to input a customized cohort using therestricted_cohortargument (see Section 3.4 for details). For example, the episodes of care and readmission values in thederived_variablestable are meant to reflect scenarios where acute hospitalizations involving a medical/SCU touchpoint were subsequently followed by a readmission involving another medical/SCU touchpoint (while ignoring potentially scheduled admissions to other in-patient services like surgery). Therefore, indrm_cleandb_v4/H4H_template_v5or newer, the epicare/readmission values were derived usingrestricted_cohort = rbind(all_med, ipscu), capturing all encounters withderived_variables$all_med == TRUEor an entry in theipscutable.Return readmission encounters: By default, the
readmission()function only returns TRUE/FALSE based on whether thegenc_idwas followed by a subsequent readmission. There may be scenarios where the readmission encounter itself is of interest to the user (e.g., to analyze the exact time-to-readmission or describe diagnoses/clinical outcomes associated with the readmission encounter). In this case, the user can set thereturn_readmit_encargument to TRUE to return readmissiongenc_ids(1 additional column per readmission window) if there exists a readmission in that time window.
Readmission: CIHI flags
CIHI readmission rate calculation
There are a total of 8 CIHI flags as function input arguments:
elective_admit,death,MAID,palliative,chemo,mental,obstetric,signout. Each flag removes episodes using different criteria when set toTRUE. More details on inclusion and exclusion criteria are discussed in the section below and on the CIHI website.-
Default definition: By default, the
elective_admitanddeathflags are set to TRUE in the readmission function, while the remaining flags are set to FALSE.- Elective admissions are excluded since they are planned. Thus, episodes of care with an elective admission are not counted as true readmissions and are therefore removed from the numerator.
- Episodes of care ending in death cannot be followed by a readmission. These episodes are thus ineligible for readmission calculations and are therefore removed from the denominator.
CIHI definition: To compute readmission rates adhering to the CIHI definition, ALL 8 CIHI flags need to be set to TRUE.
Custom definition: Users may choose to set any combination of flags in the readmission function to TRUE or FALSE based on their project-specific needs. This is only recommended for users who have a thorough understanding of all flags and how they affect readmission calculations (see details below).
# example code illustrating how to obtain readmission rates in line with CIHI definitions
res <- readmission(db, elective_admit = T, death = T, MAID = T, palliative = T, chemo = T, mental = T, obstetric = T, signout = T)
readmission_7d_derived_cihi <- mean(res$readmit7, na.rm = T)
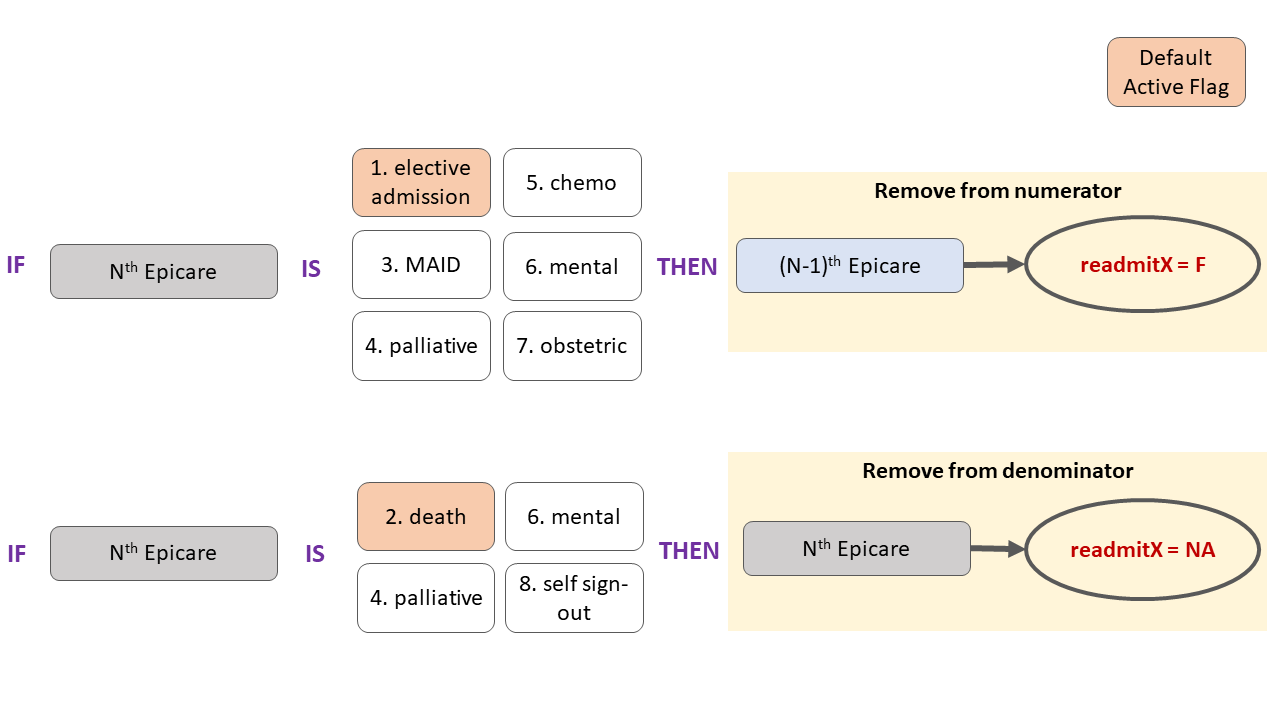
readmission_30d_derived_cihi <- mean(res$readmit30, na.rm = T)Overview of numerator & denominator removal by flag
There are 8 CIHI flags, each of which can be set to
TRUEorFALSE. Theelectiveanddeathflags areTRUEby default. Each flag results in removal of a given episode of care from the numerator and/or denominator as illustrated in the flow chart below. Note that some flags (mental & palliative) result in the removal from both numerator and denominator:
Flag details
Note: All the examples below are mock outputs of
readmission.R
Elective flag
- An episode of care is considered elective if the
FIRST encounter of that episode is elective
(
admit_category = 'L') - Episodes with elective admission are removed from the numerator because they are planned hospitalizations, and therefore, are not reflective of quality of care
Death flag
- Episodes of care ending in death are indicated by DAD Discharge Disposition Codes 07, 72, 73, and 74
- If the index admission results in death, there cannot be any
readmission. Therefore, episodes of care with death are excluded from
the denominator (
reamitX = NA)
MAID flag
- Rationale for exclusion: MAID (medical assistance in dying) is planned, and therefore, readmissions involving MAID are not reflective of quality of care
- After 2018-04-01: An episode is MAID if its discharge disposition = 73
- Before 2018-04-01: An episode is MAID if ANY
genc_idof an epicare has discharge disposition = 7 AND intervention codes indicating administration of a muscle relaxant, sedative hypnotic, and anesthetic agent. - If an epicare has ANY encounter with MAID, it is not considered to be a true readmission and is removed from the numerator.
- Note: If
death = TRUE, the epicare with MAID (which results in death) is additionally removed from the denominator. Ifdeath = FALSE, the epicare with MAID is kept in the denominator. The general recommendation is to set death toTRUEif MAID isTRUE.

Self sign-out / LAMA flag
- Rationale for exclusion: Self sign-out does not reflect physicians’ decisions to discharge, and therefore, readmission should not be attributed to discharging physician/hospital
- When LAST encounter of episode of care has discharge disposition = self sign-out (i.e. LAMA - left against medical advice: 61, 62, 65, 66, 67), the episode is removed from denominator to avoid attribution of potential readmission to physician
- Note: While self sign-out is excluded based on the CIHI definition, coding practices of self sign-out may vary widely across hospitals/physicians (e.g., some physicians may not record self sign-out since it is a patient’s right to leave on their own). Please keep that in mind if your analyses involve comparisons of readmission rates between providers.
Palliative flag
- Rationale for exclusion: Palliative care is not considered acute
care, and therefore, readmissions involving palliative care are not
reflective of quality of care; additionally, encounters with palliative
care typically result in death, and thus, the patient is not eligible
for readmission calculations (see
deathflag above) - An episode is palliative care if it has an ICD-10-CA diagnosis code of Z51.5 as type MRDx
- When an episode of care includes ANY encounter with palliative care as most responsible discharge diagnosis, it is removed from both the numerator and denominator
Chemo flag
- Rationale for exclusion: Chemotherapy is planned, and therefore, any readmissions involving chemotherapy are not reflective of quality of care
- An episode has chemotherapy if it has an ICD-10-CA diagnosis code of Z51.1 as type M, 1, C, W, X or Y
- When an episode of care includes ANY encounter with chemotherapy, the entire epicare is considered as a chemotherapy episode.
- Readmission with chemotherapy is not considered a true readmission, and it is removed from numerator.
Mental health flag
- Rationale for exclusion: Admissions for mental health diagnoses are not considered acute care
- An episode is mental health if it falls under MCC = 17
- When an episode of care includes ANY encounter with a mental health diagnosis, it is removed from numerator and denominator
Obstetric delivery
- Rationale for exclusion: Admissions for obstetric delivery are planned
- An episode is obstetric delivery if it has an ICD-10-CA diagnosis codes of: O10–O16, O21–O29, O30–O37, O40–O46, O48, O60–O69, O70–O75, O85–O89, O90–O92, O95, O98, O99 with a sixth digit of 1 or 2; or Z37 recorded in any diagnosis field
- Episodes of care that have ANY encounter with obstetric delivery are not considered a true readmission, and are therefore removed from numerator
Practical examples & special scenarios
This flow chart provides an overview of how readmission calculations can be customized for different research purposes. For many research projects, users can simply use the derived readmission variables in the DB (see Section 3.1). However, if you need further customization, you can run the readmission function with customized CIHI flags (Section 3.2), readmission window (Section 3.3), and/or a restricted cohort (Section 3.4):
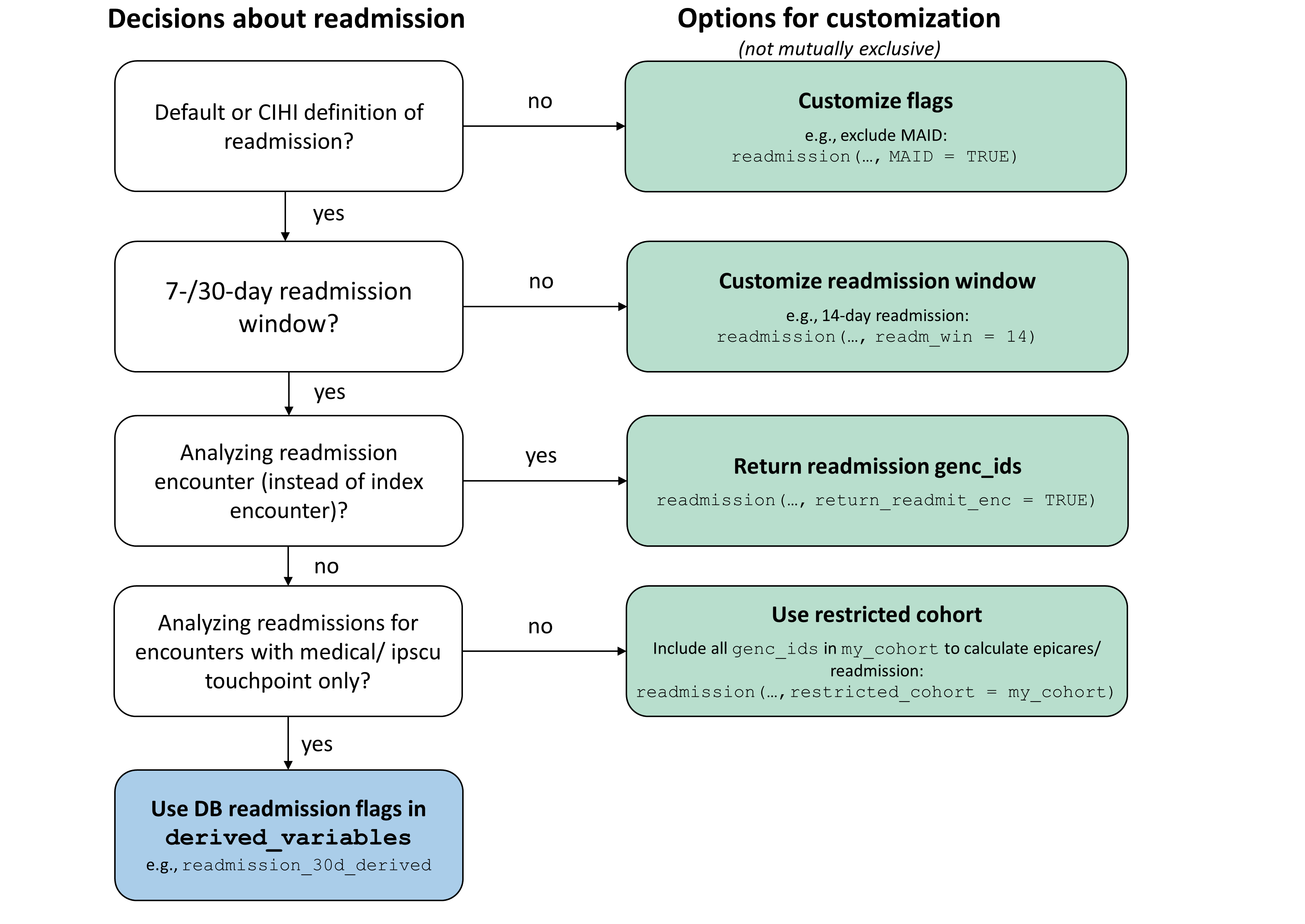
Using derived variables in DB
When to use:
- To get default/CIHI definition of 7-/30-day readmission, including all readmissions to any GEMINI hospital for all encounters with a medical/SCU touchpoint*
- The derived flags are used to calculate readmission rates in GeMQIN reports and most research projects focusing on overall readmission rates for patient cohorts of interest
- *Note that encounters without any medical/SCU touchpoint are
excluded from the derived variables (i.e.,
epicare = NAandreadmission = NA)
How to use: Query the derived_variables
table, which contains the two most commonly used definitions of 7- and
30-day readmission:
- Default definition of readmission (excluding elective readmissions
and death):
readmission_7d_derived/readmission_30d_derived - CIHI definition of readmission (all exclusion flags, see Section 2):
readmission_7d_derived_cihi/readmission_30d_derived_cihi
Example: 7-day readmission rates of patients at a given hospital:
# get derived readmissions for hospital_num 100
readm <- dbGetQuery(
db, "SELECT readmission_7d_derived FROM derived_variables WHERE hospital_num = 100;"
)
mean(readm$readmission_7d_derived, na.rm = T)Customizing CIHI flags
If projects require a customized definition of readmission rates, users can set each of the 8 CIHI flags to either TRUE or FALSE (see Section 2 for details). For example, run this code to exclude MAID (in addition to excluding elective admissions & death which are set to TRUE by default):
readmission(db, MAID = T)Specifying custom readmission window
By default, the readmission function only calculates 7-
and 30-day readmission. Users can provide an additional input argument
readm_win that contains any readmission window(s) of
interest (in days).
Example: Get 14-, 90-, and 180- day readmission.
# specify readmission windows in days
readm <- readmission(db, readm_win = c(14, 90, 180))Note: Large readmission windows can lead to the exclusion of a high percentage of encounters at the end of the data availability timeline. This is because the function can only calculate readmissions for patients whose index encounter was prior to the buffer period. The buffer period depends on the length of the readmission window (e.g., 14/90/180 days) + an additional 30 days (see Section 1.2.2). Therefore, the buffer period is longer for large readmission windows, which can lead to the exclusion of many patients. This is especially problematic if the cohort of interest focuses on the most recent data available in GEMINI (e.g., last year of available data).
Running readmission function with
restricted_cohort
You can run the readmission function on a subset of
encounters in the GEMINI database by providing an optional
restricted_cohort input.
When to use:
- To calculate readmissions within specific hospitals/subservices (e.g., readmission from and to GIM only)
- To include/exclude certain encounters from readmission calculation due to research context (e.g., readmission involving a specific diagnosis)
Important: Running the readmission
function with restricted_cohort means that only the
encounters included in the restricted cohort will be used to derive
episodes of care and readmissions. All other encounters are excluded
from all computations.
As mentioned above, the readmission flags in the
derived_variables table were calculated with
restricted_cohort = rbind(all_med, ipscu) to obtain
readmissions from/to services providing acute care (all-Med/SCU).
Similarly, if your research focuses specifically on surgery patients,
you may want to consider restricting the cohort to encounters with
surgical services only. However, note that the current
readmission function was specifically developed for acute
medical/SCU services, whereas CIHI proposes a different readmission
definition for surgical patients.
How to use:
To calculate readmission rates that only consider readmissions from
and to certain hospitals/subservices/encounters, you can run the
readmission function using a restricted_cohort, containing
all genc_ids you want to consider in the
epicare/readmission calculations.
Example 1: For example, to restrict epicare & readmission calculations to the GIM cohort:
# only consider GIM encounters in readmission calculation
readm <- readmission(db, restricted_cohort = gim_cohort)This means that only genc_ids meeting the GIM definition
(e.g., see derived_variables$gim == TRUE) will be
considered as index encounters and potential readmission encounters.
Example 2:
In certain situations, it may be meaningful to define the
restricted_cohort based on episodes of care, rather than
individual genc_ids to ensure all linked encounters are
accounted for in the readmission calculations. This may be particularly
relevant when calculating readmissions with certain diagnoses - e.g., to
accurately capture readmission rates of patients with COVID-19 who are
readmitted with a COVID-19 diagnosis. In this
case, the restricted_cohort may include all
encounters associated with episodes of care that involve at least
1 COVID-19 diagnosis (to ensure we capture linked transfers even if
a COVID-19 diagnosis was not coded for all encounters associated with
that epicare):
## Get all encounters with COVID-19 diagnosis
COVID_enc <- ipdiagnosis[grepl("^U07", diagnosis_code), "genc_id"]
## Calculate epicares based on ALL encounters in DB
# (to ensure all linked transfers are included, regardless of diagnosis codes)
epicares <- episodes_of_care(db)
## Identify all epicares that have at least 1 COVID-19 encounter
COVID_epicares <- epicares[genc_id %in% COVID_enc$genc_id, "epicare"]
## Get all encounters associated with COVID-19 epicares
# (keep all encounters of those epicares, even those without COVID diagnosis
# to make sure all linked transfers are accounted for)
COVID_epicares_all_enc <- epicares[epicare %in% COVID_epicares$epicare, "genc_id"]
## Calculate readmissions involving a COVID-19 diagnosis (considering all linked transfers in DB)
COVID_readm <- readmission(db, restricted_cohort = COVID_epicares_all_enc)
## Get 30-day readmission rate of patient with COVID-19 diagnosis
# only counting readmissions if the *readmission* epicare also involved a COVID-19 diagnosis
# if there are multiple encounters per epicare, readmission is attributed to the last encounter
# within each COVID-19 epicare
mean(COVID_readm$readmit30, na.rm = T)The resulting readmission rates indicate whether patients with
COVID-19 at the time of the index epicare had ANY subsequent readmission
for COVID-19 within a 7-/30-day time window. Those readmissions are
not necessarily the first readmission after the index
epicare! That is, there may have been intermediate epicares not
involving any COVID-19 diagnoses, which were ignored in this readmission
calculation due to the use of restricted_cohort.
Note: In the examples above, the
restricted_cohort input is only relevant because we are
restricting where patients are readmitted to
(e.g., to a specific hospital/service/epicare involving a diagnosis of
interest). If you only want to filter your cohort according to where
patients are readmitted from, you should
NOT use the restricted_cohort argument.
Instead, simply filter the output of the readmission function (or the
derived readmission flags in the database) for your encounters of
interest (see example in Section
3.1).
Other special scenarios:
There may be more complex scenarios where the inclusion criteria for
index epicares (“readmission from”) are different from the inclusion
criteria for readmission encounters (“readmission to”). For example, you
might want to analyze readmission rates of surgery patients who are
readmitted to GIM. In this case, your restricted_cohort
should include all epicares that meet the criteria for index admissions
plus any epicares that meet the criteria for readmissions. You will need
to use several steps to create your restricted_cohort:
- Select all encounters of episodes of care that meet the criteria for index admissions: In the example scenario, include all encounters of epicares that have at least one surgery encounter. If you only want to strictly consider readmissions to GIM, only include a single surgery episode of care per patient in order to avoid that subsequent surgeries are counted as readmissions.
- Select all encounters of episodes of care that meet the criteria for readmission encounters: In this example, to qualify as a GIM readmission, include all encounters from patients included in step 1 that a) occurred after the index surgery admission and b) involved an admission to GIM. Note: the implication of this step is that any intermediate non-GIM epicares are ignored.
- Combine all encounters from step 1 & 2 to create the
restricted_cohortof interest. - Run the readmission function with
restricted_cohort. - Calculate readmission rates: Filter the output of the
readmissionfunction based on your index admissions (cohort created in step 1) in order to get readmission rates of surgery patients to GIM. This step is important to exclude any readmissions following GIM (e.g., GIM to GIM readmissions or GIM to surgery readmissions) from the readmission rates.
These steps are meant as a general guideline, but you may need to
consider additional aspects for a given research project. Please make
sure you carefully consider the implications of the decisions you make
when building your restricted_cohort. You will also need to
consider if any additional REB restrictions apply, in which case the
restricted_cohort should be filtered based on REB
considerations first before applying any additional inclusion steps.
FAQ
Why are there missing values in the readmission flags?
Missing values (NA) in the returned readmission flags
indicate that readmission status for these encounters cannot be
determined or is not relevant. Encounters with NA are
excluded from the denominator in readmission rate calculations, and
thus, should be removed from any readmission analyses. There are several
reasons for NA in the readmission flags:
Buffer period: Any
genc_idsthat were discharged during the buffer period (end of data availability period for a given hospital) are returned asreadmit = NAbecause readmission cannot be reliably determined for these encounters (see here for more details). Note that longer readmission windows will lead to a longer buffer period (i.e., moreNA).Attribution to last encounter per episode of care: If there are multiple encounters in a given episode of care, readmission will only be attributed to the last encounter. All previous encounters associated with the same episode of care will be returned as
readmit = NA(see here). If attribution to the last encounter (i.e., the last discharging physician/hospital) is irrelevant for your research question, you may consider assigning the last readmission value to all encounters of a given episode of care. This could be relevant if you are only analyzing the first encounter per patient (or a randomly selected encounter).Acute transfers to non-GEMINI site: If an encounter is followed by a transfer to an acute care institution but the transfer encounter does not exist in GEMINI data (e.g., transfer to non-GEMINI hospital), all encounters of that episode of care will be returned with
readmit = NA(see here). This is because readmission is typically attributed to the last encounter of an episode of care (see point 2 above), which in this case does not exist in our data. Additionally, we don’t know how long a patient was hospitalized for at a non-GEMINI site, so we cannot reliably determine if any subsequent encounter occurred within the readmission window of interest.CIHI flags: If set to
TRUE, the following CIHI flags will result inreadmit = NAfor encounters meeting the relevant criteria (see here for details):death(TRUEby default),palliative,mental, andsignoutRestricted cohort: When using a
restricted_cohortinput, any encounters outside of that cohort will havereadmit = NA. Remember that the readmission flags in thederived_variablestable were derived withrestricted_cohort = rbind(all_med, ipscu), capturing all encounters withderived_variables$all_med == TRUEor an entry in theipscutable. This means that any encounters without medical/SCU touchpoint have a missing readmission flag (see here).